Introduction
Xenobiotic metabolism is the set of metabolic pathways that modify the chemical structure of xenobiotics, such as environmental components and pharmaceuticals to endogenously produce reactive substances (Dekant, 2009; Liska et al., 2006). The biotransformation system involves several enzyme systems that are commonly divided into two phases; phase I and phase II. The phase I enzymes are responsible for oxidation, reduction or hydrolysis and can be either detoxifying or activating (Liska, 1998). The phase II enzymes exert mainly detoxifying potential by conjugation (Liska et al., 2006). Xenobiotic metabolising enzymes are critical components in removing or detoxifying reactive metabolites of xenobiotics which make these enzyme candidates risk factors (Xu et al., 2005). The outcome of biotransformation in most cases is detoxification; nevertheless, metabolism of some xenobiotics produces metabolites that are more reactive than their substrate compound. Glutathione S-transferases are members of the phase II biotransformation enzymes. They play a key role in cellular detoxification by protecting the cell through conjugation of glutathione from chemical carcinogens and xenobiotics. These glutathione conjugates are normally less toxic and possess a better water-solubility than the original substances, whereby the excretion from the cell is facilitated.
Glutathione S-transferase T1 (GSTT1) belongs to theta class of GSTs and is involved in the detoxification of carcinogens, as well as in the formation of toxic metabolites. The gene encoding GSTT1 maps on chromosome 22q11.23 (Pemble et al., 1994) and is located within a genomic region of segmental duplications (de Bustos et al., 2006). A deletion of the GSTT1-gene (GSTT1-0 allele) is associated with susceptibility to several malignancies (Cheng et al., 2012; Hayes et al., 2005; Pan et al., 2012; Ruiz-Cosano et al., 2012). Glutathione S-transferase P1 (GSTP1) belongs to theta class of GSTs enzyme and plays a key role in biotransformation and bioactivation of certain environmental pollutants. GSTP1 gene belongs to the pi class gene family. The gene encoding GSTP1 is located on chromosome 11q13 (Board et al., 1989) and is comprised of seven exons (Morrow et al., 1989). The well studied functional polymorphism is an A to G transition at nucleotide 313 in exon 5 of GSTP1 gene which results in an isoleucine to valine substitution in codon 105 (Ile105Val) which is located on the substrate-binding site of GSTP1 (Hu et al., 1997). Since the initial identification of GST polymorphisms, a large number of genetic association studies have been conducted to investigate the relationship between these polymorphisms and the risk of disease or treatment outcome (Davies et al., 2001; Mossallam et al., 2006; Qadri et al., 2011). GST gene polymorphisms have also been studied with respect to various cancers and have been variously associated with cervical cancer (Zhang et al., 2012), esophageal cancer (Yi and Li, 2012), colorectal cancer (Wang et al., 2012), and Hodgkin and non-Hodgkin lymphoma (Bin and Luo, 2013). Given the potential important role of these polymorphisms in this study, the authors examined GSTT1 (null allele) and GSTP1(Ile105Val) polymorphisms in six south Indian populations in order to understand the frequency differences between ethnic groups that most likely result from evolutionary processes.
Materials and Methods
Subjects
Intravenous blood samples (about 3mL each) were obtained from a total of 212 unrelated males (aged 34 to 60 years old) belonging to Reddy, Balija, Mala, Madiga, Sugali and Muslims populations inhabiting Andhra Pradesh, southern peninsula of India. Apart from Muslims (religious group) five population groups speak a branch of Dravidian linguistic group. Details of the populations are documented in Table 1. All study subjects were apparently healthy volunteers and no diagnosis was performed on them. Subjects with personal or family history of cancer and any other complex diseases like diabetes and hypertension were excluded from the study. All participants provided written informed consent. This study was approved by the Ethics Committee of the Narayana Medical College, Nellore, India and conforms to the principles outlined in the Declaration of Helsinki. DNA from all samples was isolated using a standard protocol (Sambrook et al., 1989).
Table 1. Distribution of GSTP1 Ile105Val and GSTT1 Insertion-Deletion Frequencies among 6 Indian Populations
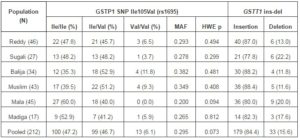
MAF; Minor Allele Frequency, HWE p; Hardy-Weinberg equilibrium p value.
Genotyping
The GSTT1 ins-del and GSTP1 Ile105Val polymorphisms were genotyped in 212 individuals. The GSTT1 genotyping was performed using PCR-electrophoresis method, to detect the presence or absence of a 215-bp product as described by Voso et al. (2002). Briefly, the primers of GSTT1 were 5’-TTC CTT ACT GGT CCT CAC ATC TC-3’ and 5’-TCA CCG GAT CAG GCC AGC A-3’. The PCR products were separated by 3% agarose gel electrophoresis and identified by ethidium bromide staining. Non-amplification upon repeated PCR denotes null allele, but this assay does not discriminate heterozygotes from homozygotes for GSTT1 null allele. The GSTP1 Ile105Val polymorphism was genotyped according to the methods previously described (Zhao et al., 2001). Briefly, the primers of GSTP1 were 5’-CCA GTG ACT GTG TGT TGA TC-3’ and 5’- CAA CCC TGG TGC AGA TGC TC-3’. The PCR products were digested with restriction enzyme BsmA I at 55℃ for 4 hours for GSTP1 genotyping and detected by electrophoresis on 3% agarose gel. For the genotyping of the GSTP1 gene, the 189-bp PCR product remained intact for the A allele, but was cleaved into smaller fragments of 149-bp and 40-bp in the case of the G allele.
Statistical Analysis
For GSTT1 ins-del polymorphism, deleted allele frequencies in each population were calculated. For GSTP1 Ile105Val polymorphism, allele frequencies in each population were determined by direct counting. Hardy-Weinberg equilibrium (HWE) ratios were calculated by software HWSIM, a DOS-based program (Cubells et al., 1997). For a worldwide comparison in a wider context the authors also extracted 20kb up and downstream SNPs around rs1695 from the HapMap data (Tanaka, 2009).
Results
The GSTT1 ins-del and GSTP1 Ile105Val were polymorphic in the study populations. Population-specific genotypes counts and frequencies among different populations are shown in Table 1. The GSTT1 (del) allele frequency varied from 11.6% in Muslims to 22.2% in Sugali populations. The very nature of GSTT1 ins-del polymorphism confined the paper performing Hardy-Weinberg equilibrium calculation. Genotype frequencies of GSTP1 Ile105Val polymorphism followed Hardy-Weinberg equation in all populations. The GSTP1 Ile105Val SNP homozygous wild type genotype (AA; Ile/Ile) frequency was 35.3% in Balija to 60.0% in Mala populations, whereas the heterozygous genotype (AG; Ile/Val) was 52.9% in Balija to 40.0% in Mala populations. The homozygous mutant genotypes (GG; Val/Val) frequencies are very low, ranging from 11.8% in Balija to 3.2% in Sugali populations, but in Mala population we could not detect this genotype. The minor allele frequency of Ile105Val SNP varied from 20.0% in Mala to 38.2% in Balija populations. The minor allele frequencies of val105 allele in different HapMap populations are documented in Table 2. The highest frequency of Val105 allele was observed in African populations (ASW, LWK, MKK and YRI) followed by European (CEU, MEX and TSI) and Indian populations (GIH). East Asian populations (CHB, CHD and JPT) showed the lowest frequency of Val105 allele. Calculation of LD in the 20kb up and downstream regions from Ile105Val (dbSNP rs1695) in HapMap populations revealed that the European populations showed larger LD followed by the East Asian populations, but in African populations LD between the SNPs of this region is very low (Figure 1).
Table 2. Minor Allele Frequencies, Observed Heterozygosities and Hardy-Weinberg Equilibrium for GSTP1 Ile105Val in HapMap Populations
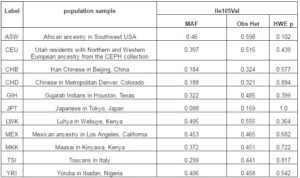 MAF; Minor allele frequency, Obs Het; observed heterozygosity, HWE p; Hardy-Weinberg equilibrium p value.
MAF; Minor allele frequency, Obs Het; observed heterozygosity, HWE p; Hardy-Weinberg equilibrium p value.
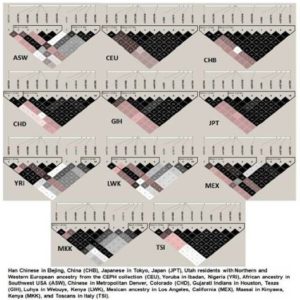
Figure 1. Linkage Disequilibrium Profiles in Different World Populations Studied in International HapMap Project. Colour Coding Represents the D’/LOD Values and the Values in Cells are r2 Multiplied by 100.
Discussion
The Glutathione S-transferases (GSTs) are the major phase II metabolizing enzymes and play a key role in cellular detoxification by the conjugation of glutathione (GSH) to a wide variety of substrates which possess electrophilic sites and convert stable and soluble compounds easily excreted from the organism (Hayes et al., 2005). In the present study, we analysed GSTT1 deletion allele that result in lack of GSTT1 activity (Sprenger et al., 2000) and GSTP1 Ile105Val polymorphism that associated with a higher level of DNA adducts (Ryberg et al., 1997). Genotypic screening of these two polymorphisms in South Indian populations revealed high variation in their allele frequency spectrum.
The incidence of the GSTT1 gene null allele differs among global populations. Significant differences in GSTT1 null allele frequencies were observed between Caucasian, Asian, African and African American populations (Lee et al., 2008). The prevalence of GSTT1 null allele in the present study ranges from 11% to 22%, which is almost similar to the frequencies reported in Caucasians (Blackburn et al., 2006; Johansson et al., 1998; Mannervik et al., 2005). Korean population showed higher frequency of (45.3%) of GSTT1 null allele compared with the white Americans (20.4%), African Americans (21.8%), Mexican-Americans (9.7%) (Hoglund et al., 2009; Marinkovic et al., 2008) and Turkish populations (10.8–28.3%) (Oke et al., 1998; Shchipanov et al., 2008; Sura et al., 2008). The GSTT1 null allele frequency in Native Russians is very close to allelic frequencies observed in some European populations (Baysal et al., 2008). GSTT1 null allele in Ouangolodougou, a north Ivory Coast population, is significantly higher (33.1%) than in Chinese, Japanese and Pakistani populations (Santovito et al., 2010; Shaikh et al., 2010). In the HapMap CEU population, it was demonstrated that the SNP rs2266633 (Asp141Asn) is the “tagging SNP” of the GSTT1 homozygous deletion (Zhao et al., 2009).
In fact, the prevalence of different GSTP1 genotypes varies between different populations and ethnic groups. The prevalence of GSTP1 Val105 in the present study ranges from 20% to 38.2% which is slightly higher than the frequencies reported in other Asian Populations (Harris et al., 1998; Pae et al., 2003). A Lesser frequency of GSTP1 Val allele was also reported in Somalis (8%) (Buchard et al., 2007), sub-Saharan Africans (12 to 21%)(Dandara et al., 2002) and Arabs (13%) (Buchard et al., 2007). A Slightly higher frequency of Val105 allele was found in Caucasians (Harris et al., 1998; Juronen et al., 2000; Rybicki et al., 2006). But analysis of HapMap populations revealed the highest frequency of Val105 allele in African populations followed by European and Indian populations.
Association Studies involving the GST genes provided conflicting results. For the elucidation of association studies, we can presume that individual functional polymorphisms might have the same functional effect in different racial groups because the versatile physiological function of the GST system will not be drastically different between ethnic populations. However, the present study reports the frequency of GSTT1 null allele and GSTP1 Ile105Val polymorphism in Indian populations which provide foundation for potential epidemiological and clinical studies.
Acknowledgment: Special appreciation is given to all blood donors whose DNA samples were used in the present study.
Conflict of Interest: There are no conflicts of interests.
(adsbygoogle = window.adsbygoogle || []).push({});
References
Baysal, E., Bayazit, Y. A., Ceylaner, S., Alatas, N., Donmez, B., Ceylaner, G., San, I., Korkmaz, B., Yilmaz, A., Menevse, A., Altunyay , S., Gunduz, B., Goksu, N., Arslan, A. & Ekmekci, A. (2008). “GJB2 and Mitochondrial A1555g Gene Mutations in Nonsyndromic Profound Hearing Loss and Carrier Frequencies in Healthy Individuals,” J Genet, 87 (1) 53-57.
Publisher – Google Scholar – British Library Direct
Bin, Q. & Luo, J. (2013). “Role of Polymorphisms of GSTM1, GSTT1 and GSTP1 Ile105val in Hodgkin and Non-Hodgkin Lymphoma Risk: A Human Genome Epidemiology (Huge) Review,” Leuk Lymphoma, 54 (1) 14-20.
Publisher – Google Scholar
Blackburn, A. C., Matthaei, K. I., Lim, C., Taylor, M. C., Cappello, J. Y., Hayes, J. D., Anders, M. W. & Board, P. G. (2006). “Deficiency of Glutathione Transferase Zeta Causes Oxidative Stress and Activation of Antioxidant Response Pathways,” Mol Pharmacol, 69 (2) 650-657.
Publisher – Google Scholar – British Library Direct
Board, P. G., Webb, G. C. & Coggan, M. (1989). “Isolation of a Cdna Clone and Localization of the Human Glutathione S-Transferase 3 Genes to Chromosome Bands 11q13 and 12q13-14,” Ann Hum Genet, 53 (Pt 3) 205-213.
Publisher – Google Scholar
Buchard, A., Sanchez, J. J., Dalhoff, K. & Morling, N. (2007). “Multiplex PCR Detection of GSTM1, GSTT1, and GSTP1 Gene Variants: Simultaneously Detecting GSTM1 and GSTT1 Gene Copy Number and the Allelic Status of the GSTP1 Ile105val Genetic Variant,” J Mol Diagn, 9 (5) 612-617.
Publisher – Google Scholar – British Library Direct
Cheng, H. Y., You, H. Y. & Zhou, T. B. (2012). “Relationship between GSTM1/GSTT1 Null Genotypes and Renal Cell Carcinoma Risk: A Meta-Analysis,” Ren Fail, 34 (8) 1052-1057.
Publisher – Google Scholar
Cubells, J. F., Kobayashi, K., Nagatsu, T., Kidd, K. K., Kidd, J. R., Calafell, F., Kranzler, H. R., Ichinose, H. &
Gelernter, J. (1997). “Population Genetics of a Functional Variant of the Dopamine Beta-Hydroxylase Gene (DBH),” Am J Med Genet, 74 (4) 374-379.
Publisher – Google Scholar – British Library Direct
Dandara, C., Sayi, J., Masimirembwa, C. M., Magimba, A., Kaaya, S., De Sommers, K., Snyman, J. R. & Hasler, J. A. (2002). “Genetic Polymorphism of Cytochrome P450 1a1 (CYP1A1) and Glutathione Transferases (M1, T1 and P1) among Africans,” Clin Chem Lab Med, 40 (9) 952-957.
Publisher – Google Scholar – British Library Direct
Davies, S. M., Robison, L. L., Buckley, J. D., Tjoa, T., Woods, W. G., Radloff, G. A., Ross, J. A. & Perentesis, J. P. (2001). “Glutathione S-Transferase Polymorphisms and Outcome of Chemotherapy in Childhood Acute Myeloid Leukemia,” J Clin Oncol, 19 (5) 1279-1287.
Publisher – Google Scholar – British Library Direct
De Bustos, C., Diaz de Stahl, T., Piotrowski, A., Mantripragada, K. K., Buckley, P. G., Darai, E., Hansson, C. M., Grigelionis, G., Menzel, U. & Dumanski, J. P. (2006). “Analysis of Copy Number Variation in the Normal Human Population within a Region Containing Complex Segmental Duplications on 22q11 Using High-Resolution Array-Cgh,” Genomics, 88 (2) 152-162.
Publisher – Google Scholar
Dekant, W. (2009). “The Role of Biotransformation and Bioactivation in Toxicity,” EXS, 99 57-86.
Publisher – Google Scholar
Harris, M. J., Coggan, M., Langton, L., Wilson, S. R. & Board, P. G. (1998). “Polymorphism of the Pi Class Glutathione S-Transferase in Normal Populations and Cancer Patients,” Pharmacogenetics, 8 (1) 27-31.
Publisher – Google Scholar – British Library Direct
Hayes, J. D., Flanagan, J. U. & Jowsey, I. R. (2005). “Glutathione Transferases,” Annu Rev Pharmacol Toxicol, 45 51-88.
Publisher – Google Scholar – British Library Direct
Hoglund, J., Gustafsson, K., Ljungstrom, B. L., Engstrom, A., Donnan, A. & Skuce, P. (2009). “Anthelmintic Resistance in Swedish Sheep Flocks Based on a Comparison of the Results from the Faecal Egg Count Reduction Test and Resistant Allele Frequencies of the Beta-Tubulin Gene,” Vet Parasitol, 161 (1-2) 60-68.
Publisher – Google Scholar
Hu, X., Xia, H., Srivastava, S. K., Herzog, C., Awasthi, Y. C., Ji, X., Zimniak, P. & Singh, S. V. (1997). “Activity of Four Allelic Forms of Glutathione S-Transferase Hgstp1-1 for Diol Epoxides of Polycyclic Aromatic Hydrocarbons,” Biochem Biophys Res Commun, 238 (2) 397-402.
Publisher – Google Scholar – British Library Direct
Johansson, A.- S., Stenberg, G., Widersten, M. & Mannervik, B. (1998). “Structure-Activity Relationships and Thermal Stability of Human Glutathione Transferase P1-1 Governed by the H-Site Residue 105,” J Mol Biol, 278 (3) 687-698.
Publisher – Google Scholar – British Library Direct
Juronen, E., Tasa, G., Veromann, S., Parts, L., Tiidla, A., Pulges, R., Panov, A., Soovere, L., Koka, K. &
Mikelsaar, A. -V. (2000). “Polymorphic Glutathione S-Transferases as Genetic Risk Factors for Senile Cortical Cataract in Estonians,” Invest Ophthalmol Vis Sci, 41 (8) 2262-2267.
Publisher – Google Scholar – British Library Direct
Lee, M. Y., Mukherjee, N., Pakstis, A. J., Khaliq, S., Mohyuddin, A., Mehdi, S. Q., Speed, W. C., Kidd, J. R. &
Kidd, K. K. (2008). “Global Patterns of Variation in Allele and Haplotype Frequencies and Linkage Disequilibrium across the CYP2E1 Gene,” Pharmacogenomics J, 8 (5) 349-356.
Publisher – Google Scholar
Liska, D. J. (1998). “The Detoxification Enzyme Systems,” Altern Med Rev, 3 (3) 187-198.
Publisher – Google Scholar
Liska, D., Lyon, M. & Jones, D. S. (2006). “Detoxification and Biotransformational Imbalances,” Explore (NY), 2 (2) 122-140.
Publisher – Google Scholar
Mannervik, B., Board, P. G., Hayes, J. D., Listowsky, I. & Pearson, W. R. (2005). “Nomenclature for Mammalian Soluble Glutathione Transferases,” Methods Enzymol, 401 1-8.
Publisher – Google Scholar – British Library Direct
Marinkovic, N., Pasalic, D., Grskovic, B., Ferencak, G., Honovic, L. & Rukavina, A. S. (2008). “Genotype Frequencies of Udp-Glucuronosyltransferase 1a1 Promoter Gene Polymorphism in the Population of Healthy Croatian Pre-Scholars,” Coll Antropol, 32 (3) 725-729.
Publisher – Google Scholar
Morrow, C. S., Cowan, K. H. & Goldsmith, M. E. (1989). “Structure of the Human Genomic Glutathione S-Transferase-Pi Gene,” Gene, 75 (1) 3-11.
Publisher – Google Scholar
Mossallam, G. I., Abdel Hamid, T. M. & Samra, M. A. (2006). “Glutathione S-Transferase GSTM1 and GSTT1 Polymorphisms in Adult Acute Myeloid Leukemia; its Impact on Toxicity and Response to Chemotherapy,” J Egypt Natl Canc Inst, 18 (3) 264-273.
Publisher – Google Scholar
Oke, B., Akbas, F., Aydin, M. & Berkkan, H. (1998). “GSTT1 Null Genotype Frequency in a Turkish Population,” Arch Toxicol, 72 (7) 454-455.
Publisher – Google Scholar – British Library Direct
Pae, C. U., Kim, J. J., Lee, S. J., Lee, C. U., Lee, C., Paik, I. H., Park, H. R., Yang, S. & Serretti, A. (2003). “Association Study between Glutathione S-Transferase P1 Polymorphism and Schizophrenia in the Korean Population,” Prog Neuropsychopharmacol Biol Psychiatry, 27 (3) 519-523.
Publisher – Google Scholar
Pan, Z.- J., Huang, W.- J., Zou, Z.- H. & Gao, X.- C. (2012). “The GSTT1 Null Genotype Contributes to Increased Risk of Prostate Cancer in Asians: A Meta-Analysis,” Asian Pac J Cancer Prev, 13 (6) 2635-2638.
Publisher – Google Scholar
Pemble, S., Schroeder, K. R., Spencer, S. R., Meyer, D. J., Hallier, E., Bolt, H. M., Ketterer, B. & Taylor, J. B. (1994). “Human Glutathione S-Transferase Theta (GSTT1): Cdna Cloning and the Characterization of a Genetic Polymorphism,” Biochem J, 300 ( Pt 1) 271-276.
Publisher – Google Scholar – British Library Direct
Qadri, Q., Sameer, A. S., Shah, Z. A., Hamid, A., Alam, S., Manzoor, S. & Siddiqi, M. A. (2011). “Genetic Polymorphism of the Glutathione-S-Transferase P1 Gene (GSTP1) and Susceptibility to Prostate Cancer in the Kashmiri Population,” Genet Mol Res, 10 (4) 3038-3045.
Publisher – Google Scholar
Ruiz-Cosano, J., Conesa-Zamora, P., Gonzalez-Conejero, R., Perez-Ceballos, E., Martinez-Frances, A., Vicente, V. & Perez-Guillermo, M. (2012). “Role of GSTT1 and M1 Null Genotypes as Risk Factors for B-Cell Lymphoma: Influence of Geographical Factors and Occupational Exposure,” Mol Carcinog, 51 (6) 508-513.
Publisher – Google Scholar
Ryberg, D., Skaug, V., Hewer, A., Phillips, D. H., Harries, L. W., Wolf, C. R., Ogreid, D., Ulvik, A., Vu, P. & Haugen, A. (1997). “Genotypes of Glutathione Transferase M1 and P1 and Their Significance for Lung DNA Adduct Levels and Cancer Risk,” Carcinogenesis, 18 (7) 1285-1289.
Publisher – Google Scholar – British Library Direct
Rybicki, B. A., Neslund-Dudas, C., Nock, N. L., Schultz, L. R., Eklund, L., Rosbolt, J., Bock, C. H. & Monaghan, K. G. (2006). “Prostate Cancer Risk from Occupational Exposure to Polycyclic Aromatic Hydrocarbons Interacting with the GSTP1 Ile105val Polymorphism,” Cancer Detect Prev, 30 (5) 412-422.
Publisher – Google Scholar
Sambrook, J., Fitsch, E. F. & Maniatis, T. (1989). ‘Molecular Cloning: A Laboratory Manual,’ Cold Spring Harbor: Cold Spring Harbor Press.
Google Scholar
Santovito, A., Burgarello, C., Cervella, P. & Delpero, M. (2010). “Polymorphisms of Cytochrome P450 1a1, Glutathione S-Transferases M1 and T1 Genes in Ouangolodougou (Northern Ivory Coast),” Genet Mol Biol, 33 (3) 434-437.
Publisher – Google Scholar
Shaikh, R. S., Amir, M., Masood, A. I., Sohail, A., Athar, H. U., Siraj, S., Ali, M. & Anjam, M. S. (2010). “Frequency Distribution of GSTM1 and GSTT1 Null Allele in Pakistani Population and Risk of Disease Incidence,” Environ Toxicol Pharmacol, 30 (1) 76-79.
Publisher – Google Scholar
Shchipanov, N. A., Bulatova, N. & Pavlova, S. V. (2008). “Distribution of Two Chromosome Races of the Common Shrew (Sorex Araneus L.) in the Hybrid Zone: May a Change of the Dispersal Mode Maintain Independent Gene Frequencies?,” Genetika, 44 (6) 734-745.
Publisher – Google Scholar – British Library Direct
Sprenger, R., Schlagenhaufer, R., Kerb, R., Bruhn, C., Brockmoller, J., Roots, I. & Brinkmann, U. (2000). “Characterization of the Glutathione S-Transferase GSTT1 Deletion: Discrimination of All Genotypes by Polymerase Chain Reaction Indicates a Trimodular Genotype-Phenotype Correlation,” Pharmacogenetics, 10 (6) 557-565.
Publisher – Google Scholar – British Library Direct
Sura, T., Eu-ahsunthornwattana, J., Pingsuthiwong, S. & Busabaratana, M. (2008). “Sensitivity and Frequencies of Dystrophin Gene Mutations in Thai Dmd/Bmd Patients as Detected by Multiplex PCR,” Dis Markers, 25 (2) 115-121.
Publisher – Google Scholar
Tanaka, T. (2009). ‘Hapmap Project,’ Nihon Rinsho, 67 (6) 1068-1071.
Voso, M. T., D’Alo, F., Putzulu, R., Mele, L., Scardocci, A., Chiusolo, P., Latagliata, R., Lo-Coco, F., Rutella, S., Pagano, L., Hohaus, S. & Leone, G. (2002). “Negative Prognostic Value of Glutathione S-Transferase (GSTM1 and GSTT1) Deletions in Adult Acute Myeloid Leukemia,” Blood, 100 (8) 2703-2707.
Publisher – Google Scholar – British Library Direct
Wang, D., Zhang, L. M., Zhai, J. X. & Liu, D. W. (2012). “GSTM1 and GSTT1 Polymorphisms and Colorectal Cancer Risk in Chinese Population: A Meta-Analysis,” Int J Colorectal Dis, 27 (7) 901-909.
Publisher – Google Scholar
Xu, C., Li, C. Y. T. & Kong, A. N. T. (2005). “Induction of Phase I, Ii and Iii Drug Metabolism/Transport by Xenobiotics,” Arch Pharm Res, 28 (3) 249-268.
Publisher – Google Scholar
Yi, S. M. & Li, G. Y. (2012). “Null Genotype of GSTT1 Contributes to Esophageal Cancer Risk in Asian Populations: Evidence from a Meta-Analysis,” Asian Pac J Cancer Prev, 13 (10) 4967-4971.
Publisher – Google Scholar
Zhang, Z. Y., Jin, X. Y., Wu, R., Wu, L. N., Xing, R., Yang, S. J. & Xie, Y. (2012). “Meta-Analysis of the Association between GSTM1 and GSTT1 Gene Polymorphisms and Cervical Cancer,” Asian Pac J Cancer Prev, 13 (3) 815-819.
Publisher – Google Scholar
Zhao, M., Lewis, R., Gustafson, D. R., Wen, W.- Q., Cerhan, J. R. & Zheng, W. (2001). “No Apparent Association of GSTP1 A313G Polymorphism with Breast Cancer Risk among Postmenopausal Iowa Women,” Cancer Epidemiol Biomarkers Prev, 10 (12) 1301-1302.
Publisher – Google Scholar – British Library Direct
Zhao, Y., Marotta, M., Eichler, E. E., Eng, C. & Tanaka, H. (2009). “Linkage Disequilibrium between Two High-Frequency Deletion Polymorphisms: Implications for Association Studies Involving the Glutathione-S Transferase (GST) Genes,” PLoS Genet, 5 (5) e1000472.
Publisher – Google Scholar